All about the Nobel prize winning genome editing tool – CRISPR
- M Subha Maheshwari
- Nov 14, 2020
- 5 min read
The 2020 Nobel Prize in Chemistry was awarded to Emmanuelle Charpentier and Jennifer A. Doudna “for the development of a method for genome editing.” That method, formally known as CRISPR-Cas9 gene editing but often called simply CRISPR.
What is CRISPR?
CRISPR-Cas9 system is naturally present in the archaea and bacteria to protect from invading viruses and plasmids. It works through complementary RNA guided cleavage of the foreign DNA, which subsequently is repaired through either imprecise non-homologous end joining or template-dependent homologous directed repair systems.
History
These days, CRISPR is omnipresent in the world of gene editing. It was not always the first method of modifying a genome. The earliest methods came from the discovery of restriction enzymes and meganucleases. The model of precise genome editing was further advanced with the development of zinc-finger nucleases, a tool for gene editing. The outcome from ZFN use illustrated that it showed off-target effects. ZFNs were tricky to engineer, making them costly for the researchers and a time-taking process for the manufacturers.
Later on, transcription activator-like effector nucleases (TALENs) were developed. TALENs were more practical than ZFNs because of their DNA-binding domains. This has more potential target sequences which meant that they can now be used for more applications. TALENs were definitely a leap forward from the ZFNs, but they were too costly to manufacture despite being easier to design than ZFNs.
There is another gene editing technique that uses restriction enzymes along with recombinant adeno-associated viruses. rAAVs were limited by their small vector capacities of genetic material to be modified. Henceforth, their resulting efficiency was limited. Their vectors were also complicated to create.
CRISPR, on the other hand, is based on DNA-RNA interactions, which are commonly understood, and therefore, provides an easier, simpler way to edit genes. This simplicity and effectiveness has changed the field of gene editing forever. Compared to other gene-editing tools such as ZNS (zinc finger nucleases) and TALENS (transcription activator-like effector nucleases), CRISPR-Cas9 was cost-effective, reliable, robust, precise and multiplexed gene-editing tool.
Components of CRISPR-Cas9
CRISPR has two components – a guide RNA (gRNA) and a CRISPR-associated endonuclease (Cas). The gRNA is specific to the target DNA sequence. In experiments using CRISPR, the gRNA and Cas are combined and the result is a ribonucleoprotein (RNP) complex. Cas is a protein that acts to cut the target DNA while the gRNA guides it to the target DNA site. In prokaryotes, the gRNA is used to target viral DNA, but as a gene editing tool, it can be designed to target any gene site in almost any location. Cas9 is a popular choice of Cas proteins. Cas9 is from Streptococcus pyogenes. The gRNA finds the target site and binds to the DNA, but this binding requires the presence of a PAM (protospacer adjacent motif) immediately downstream of the target but on the opposite DNA strand. Different Cas proteins (from different prokaryotic species) recognize different PAM sequences. Cas9 is popular because of the frequency and flexibility of its PAM, which is 5’-NGG-3’. The N represents any nucleotide. This means that any DNA sequence with two G (guanine) bases can be used to form a PAM for Cas9. If the gRNA successfully binds to the target, the Cas9 cleaves both DNA strands. This cleaving process takes place 3 to 4 nucleotides upstream of the PAM site.
Guide RNA
In nature, gRNA is made of two separate sections of RNA. These two sections are CRISPR RNA (crRNA) and transactivating CRISPR RNA (tracrRNA). crRNA is 18-20 base pairs in length and binds to the DNA target sequence. TracrRNA acts as a structure which the crRNA-Cas9 interaction can take place on. In nature, the gRNA is a duplex molecule, with crRNA and tracrRNA annealed together. Synthetically, gRNA can be produced with these two molecules connected by a linker loop. These are called single guide RNAs (sgRNAs).
Creating Double-Strand Breaks
Cas9 carries out its role in CRISPR-Cas gene editing experiments by causing a DSB to form in the DNA. This resembles the process in prokaryotes. The DSB formation is the first step in the CRISPR editing process. It is followed by a repair mechanism which decides what type of gene editing will occur. Two types of repair, namely non-homologous end joining (NHEJ) and homology-directed repair (HDR).

Single guide RNAs and two component RNAs. The crRNA and tracrRNA components can be anneal together to form the two component gRNA, or the two components can be joined by a linker loop to create a continuous molecule (sgRNA).
Double Strand Break Repair
CRISPR causes DSBs at desired sites in the genome. This is just the first step in the editing process. It is the repair mechanisms that follow this that allow the editing to occur. Innate DNA repair processes automatically respond to the formation of the DSB. The two main types of repair mechanisms that are used to edit genes are non-homologous end joining (NHEJ) and homology-directed repair (HDR).
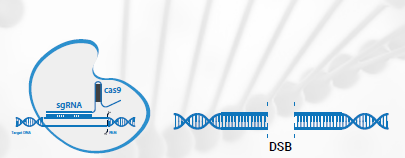
CRISPR-Cas9 produces a double strand break (DSB). Cas9 and sgRNA combine to create a ribonucleoprotein. The sgRNA binds to the target DNA and Cas9 cleaves the DNA, causing a DSB.
Non-Homologous End Joining (NHEJ)
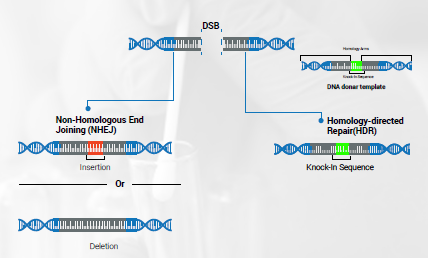
NHEJ can be used when the desired result is to permanently knockout a gene (so that no functional protein is made). NHEJ facilitates the re-joining of the DNA ends. However, it often allows erroneous changes to occur, which may result in inserted or deleted nucleotides that are not intended. These are called indels. If the numbers of nucleotide changes (inserted or deleted) are not a multiple of three, a frameshift mutation will occur. This mutation will likely eliminate the functionality of the resulting protein.
Homology Directed Repair (HDR)
If the gene editing result desired by the researchers is to replace the targeted DNA sequence with another sequence, then HDR can be used. A DNA template from a donor that possessed that desired DNA segment is introduced. This template is surrounded by sections of homologous DNA sequences. The host’s repair mechanisms will use this template to fix the DSB by using homologous recombination. By this process, the donor’s sequence is incorporated into the sequence being repaired.
Applications of CRISPR
The field of gene editing has grown vastly since CRISPR was developed. CRISPR can be used in a very large variety of cells in many different organisms, such as mammals, plants and fungi. It has been used to alter genomes in many ways, including changes in nucleotide sequences and changing the expression of genes.
Knockouts
A knockout refers to the process of making a gene permanently inactive, which means that it cannot encode a functioning protein. CRISPR can cause this because of how error-prone NHEJ can be. NHEJ often produces indels that can result in a frameshift mutation and inactivate the function of the gene. A particularly susceptible DNA sequence is a premature stop codon. Knockouts are also results of using multi-guide sgRNAs. These are gRNAs that target the same gene. The subsequent multiple cuts in the DNA induces at least one large fragment deletion in the target DNA. These deletions cause the loss of several amino acids, which will likely result in the complete inactivation of the target gene. The resulting changes in protein expression following a knockout can provide researchers with an insight into the phenotype of a cell or organism. This is useful for many applications, such as identifying and validating potential drug targets, analyzing cellular mechanisms and evaluating antibody activity.
Knock-ins
Knocking-in is the process of incorporating genetic material into a host genome. This process is facilitated by HDR, as discussed previously. In experiments using HDR, a DNA template is introduced to allow the HDR mechanism to introduce the desired change into the host genome. HDR, as a gene editing tool, allows for many applications. Examples range from single point mutation alterations to the addition of selectable markers. The HDR mechanism still requires further research to improve efficiency, but researchers have managed to use it to fix a mutation which causes cataracts in mice. This illustrates that HDR is a viable concept for correcting genetic diseases.
References
Martina Fabricci. (2020). CRISPR-Cas9. CRISPR Biotech Engineering - 'Tomorrow's Technology for Today's Challenges‘. CRISPR-Cas9. Retrieved from the World Wide Web, https://crispr-bte.com/crispr-cas9/ .
Pinkert, Carl. (2014). Transgenic Animal Technology: A Laboratory Handbook: Third Edition. 1-696.
Martina Fabricci. (2020).CRISPR Biotech Engineering - 'Tomorrow's Technology for Today's Challenges‘. Retrieved from the World Wide Web, https://crispr-bte.com/wp-content/uploads/2020/04/CRISPR-Ebook.pdf .




Comments